
| PMC full text: |
|
Figure 3.
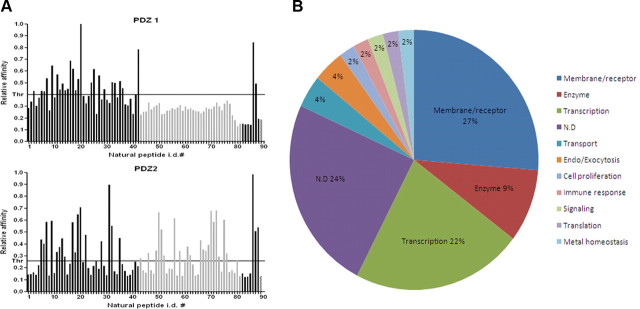
Novel putative interaction partners of PDZ domains of Mint1. A, Histograms showing the binding profile of 89 C-terminal proteomic fragments (natural ligands) to both PDZ domains. Axis X indicates identification (i.d.) numbers of individual natural ligands. Axis Y indicates the normalized relative affinity. The last four bars on the histogram indicate control peptides. Positive controls: positions 87 and 88 (ASYCKQRNIVWL and GHLSESSQVRWL, respectively) are synthetic artificial peptides isolated from a phage-displayed library, which showed relatively high affinity for both PDZ domains; position 89 is copper chaperone for superoxide dismutase (CCS) (GRKESAQPPAHL). Negative control: position 90 (gridded bar), GP41 peptide (CWFSITNWLWYI). Gray-colored bars represent peptides with C-terminal Val residue. B, Pie graph showing percentage of different cellular functionalities associated with Mint1, discovered in this study.